Module 3 | Lesson 1 | Fly-CURE - Project Overview
Overview
Teaching: min
Exercises: minQuestions
What is the Fly-CURE?
Objectives
Explain how the mutant flies were created.
Understand what research has already been done and what we know to this point.
Understand how the sequencing data was obtained.
Recorded Lesson:
Module 3 | Lesson 1 | Fly-CURE - Project Overview
Fly-CURE Overview
The Fly-CURE is a consortium of diverse universities and colleges across the US teaching a Course-Based Undergraduate Research Experience (Fly-CURE). The Fly-CURE is a 10-15 week undergraduate research experience where students characterize and map a novel mutant stock of Drosophila melanogaster. Students use techniques in classical and molecular genetics as well as bioinformatics during a semester long lab course (Fig. 1).
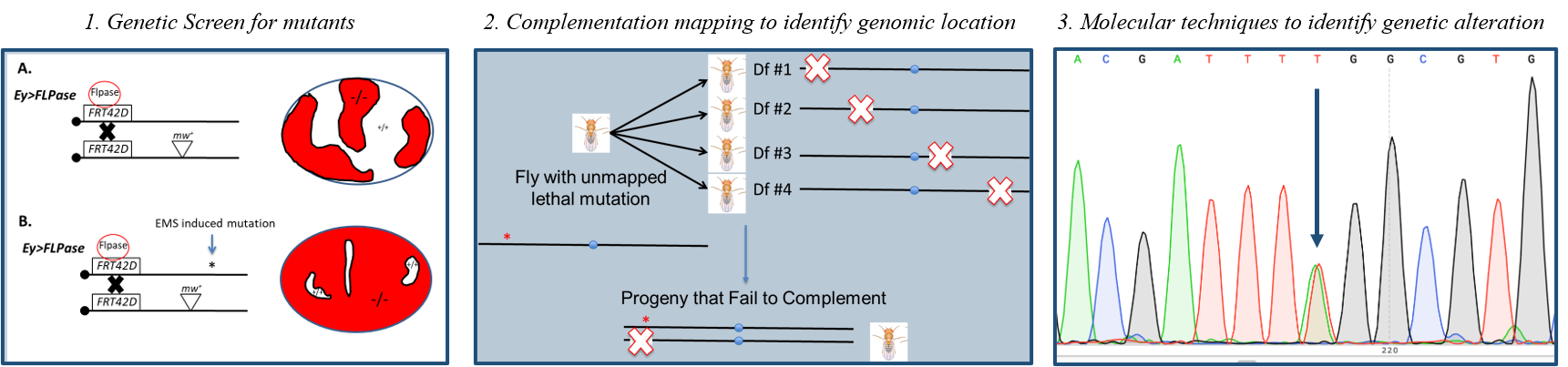
Figure 1: Overview of Fly-CURE. Step 1 includes utilizing the Ey>Flp/FRT system to screen for cell overgrowth. 1A. w;FRT42D,Dark82,/CyO X w, Ey>FLP;FRT42D 1B. w;FRT42D,Dark82,EMS/CyO X w, Ey>FLP;FRT42D. Step 2 is a model of complementation mapping conducted utilizing the Bloomington Indiana Stock Center (BDSC) 2R deficiency kit. Locations that fail to complement are further assessed by additional mapping. Step 3 includes various molecular techniques including DNA extraction, primer design, polymerase chain reaction (PCR), sanger sequencing, and bioinformatic analysis to identify a heterozygous single nucleotide polymorphism (SNP).
Genetic screen
D. melanogaster EMS mutants were screened in the background of blocked apoptosis (Dark82) to search for genetic regulators of cell growth and cell division (Fig. 2). The Fly-CURE was developed to have undergraduate researchers map these novel D. melanogaster mutants. The mutations result in homozygous recessive lethality. Heterozygotes have normal growth control and homozygous mutants are dead. In order to observe the phenotype in Fig. 2, a genetic ‘trick’ is used to induce mitotic recombination to generate homozygous mutant patches of cells only in the eye (which is not required for fly survival), while the rest of the animal remains heterozygous for the mutation. The growth control and mutant phenotype can then be studied by examining the eye tissue.
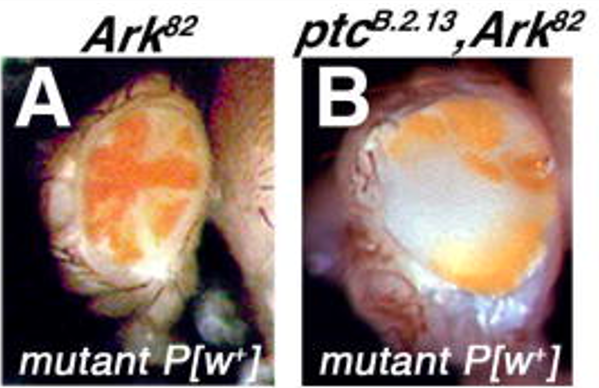
Figure 2: Example of a mosaic mutant identified in the genetic screen. Kagey et al. 2012 Mech. of Dev.
Complementation mapping and molecular analyses
Once a mutant is identified, students complete complementation mapping utilizing the Bloomington Deficiency Stock Center (BDSC) 2R deficiency kit. This kit allows for localization of the genomic location of the EMS mutant. Additional rounds of complementation mapping may be utilized to map to predicted alleles allowing for more precise primer design for the polymerase chain reaction (PCR). After PCR amplification, Sanger sequencing and bioinformatic analysis is conducted to identify the SNP generated by EMS.
Outcomes
To-date, this project has provided research exposure to over 1500 undergraduate researchers within the classroom setting. These undergraduate researchers have successfully mapped several EMS mutants which have led to local and national scientific presentations by students, as well as numerous peer-reviewed publications (Fig. 3).
- Fly-CURE, a multi-institutional CURE using Drosophila, increases students’ confidence, sense of belonging, and persistence in research
- The I.3.2 developmental mutant has a single nucleotide deletion in the gene centromere identifier
- Genetic mapping of EgfrL.3.1 in Drosophila melanogaster
- Genetic mapping of the p47L.3.2 mutation in Drosophila melanogaster
- The mapping of Drosophila melanogaster mutant A.4.4
- Genetic mapping of shnE.3.2 in Drosophila melanogaster
- Genetic mapping and phenotypic analysis of shotH.3.2 in Drosophila melanogaster
- cliffordB.4.1, an allele of CG1603 , causes tissue overgrowth in the Drosophila melanogaster eye
- B.2.16 is a non-lethal modifier of the Dark82 mosaic eye phenotype in Drosophila melanogaster
- Genetic mapping of a new Hippo allele, HpoN.1.2, in Drosophila melanogaster
- Genetic mapping of Uba3O.2.2, a pupal lethal mutation in Drosophila melanogaster
- Genetic mapping and Phenotypic analysis of GstE14E.4.1 on Eye and Antennae Development in Drosophila melanogaster
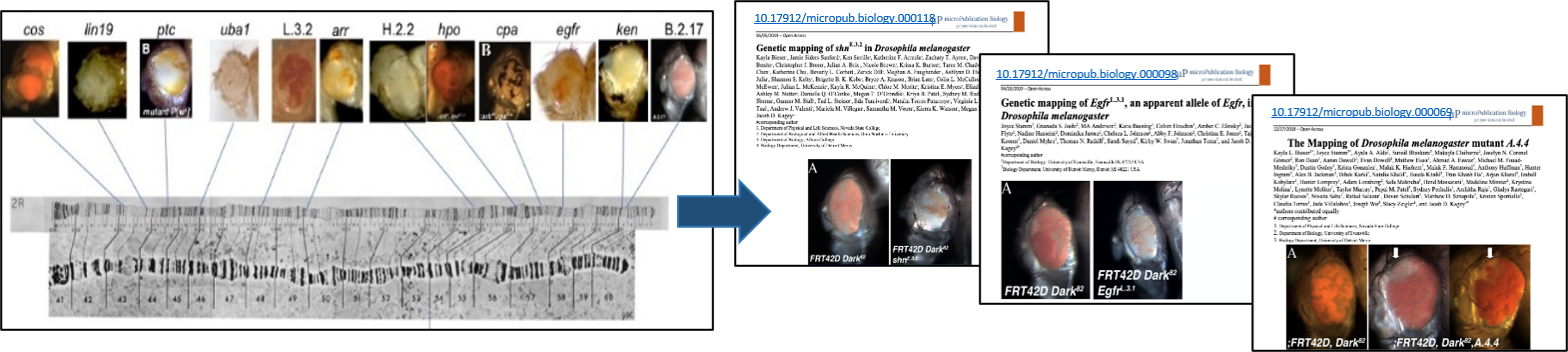
Figure 3: Mutants mapped by undergraduate researchers and sampling of Fly-CURE microPublications.
Whole-Genome Sequencing
Some EMS mutants have proven difficult to map utilizing the techniques in the Fly-CURE. As a result, numerous grants were obtained to conduct whole-genome sequencing (WGS) on the Dark82 control and many EMS mutant stocks. All mutants sequenced to-date and corresponding data can be accessed at this link.
Figure 4: Images of the Dark82 control and the 14 EMS mutants that were sequenced in the original study.
Objective
The primary objective for Module 3, is to utilize a genomics pipeline to identify the causative SNPs in EMS mutants (Fig. 5). We will be using many of the skills and tools you have previously utilized in the Module 1 (Shell Genomics) and Module 2 (Wrangling Genomics).
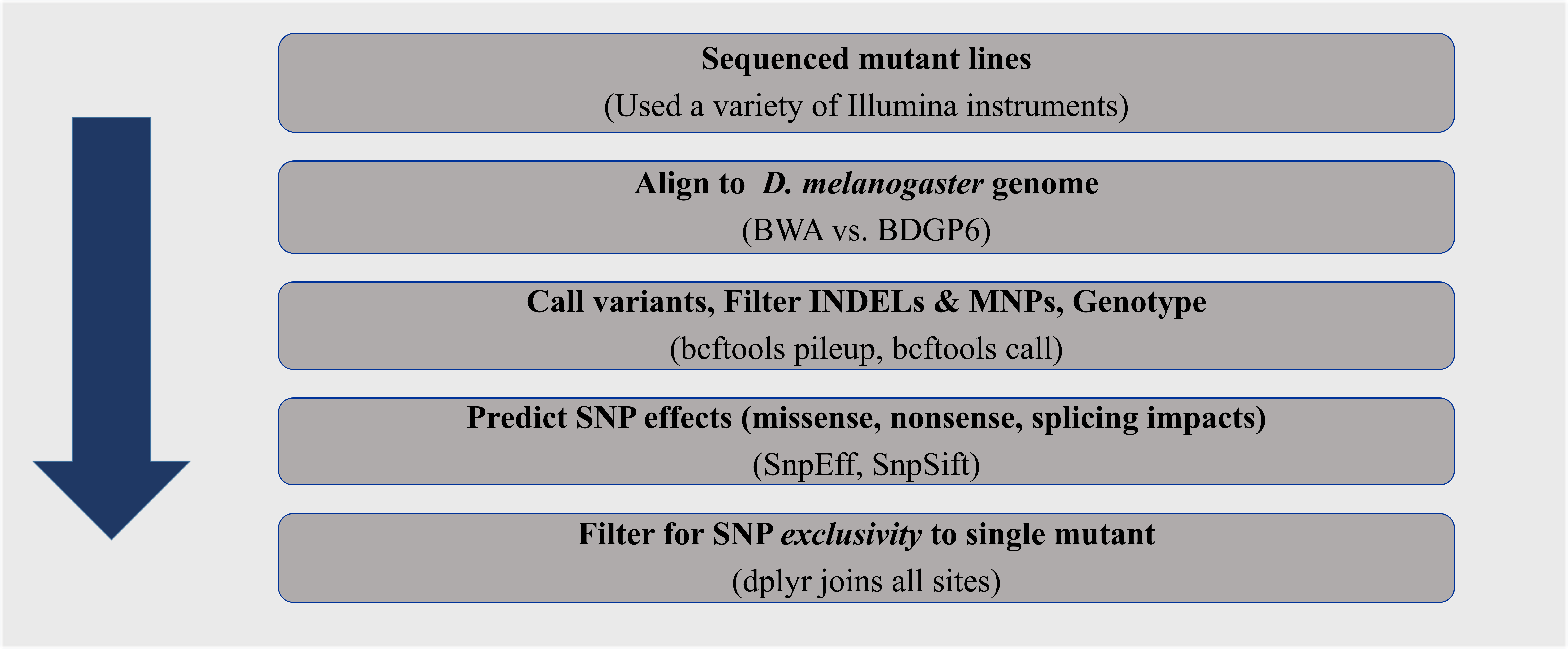
Figure 5: Genomics pipeline to identify the causative SNP in EMS Drosophila melanogaster mutants.
Funding sources
The Fly-CURE is supported by NSF IUSE # 2021146. The bioinformatics curriculum training is supported by NSF RCN # 2316218. The sequencing project described was supported by a grant from the National Institute of General Medical Sciences (GM103440).
Key Points
This is an authentic hypothesis-driven research project spanning multiple semesters and institutions.